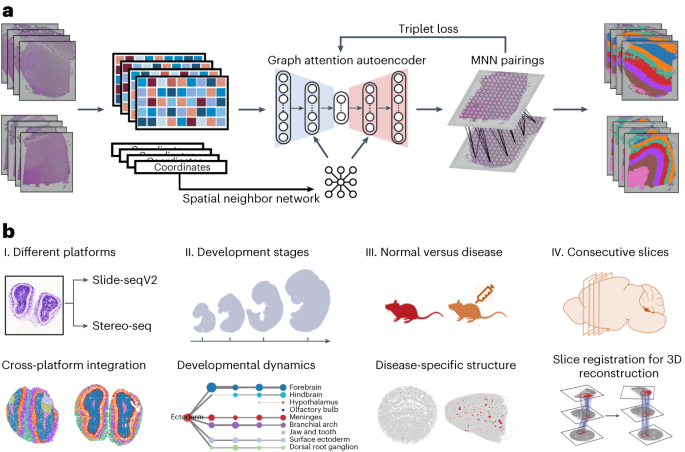
STAligner enables the integration and alignment of multiple spatial transcriptomics datasets

The first output table of the “Rosetta Tool”, showing codon (triplets), followed by single-letter translated amino acids, for each read in the input FASTA alignment.
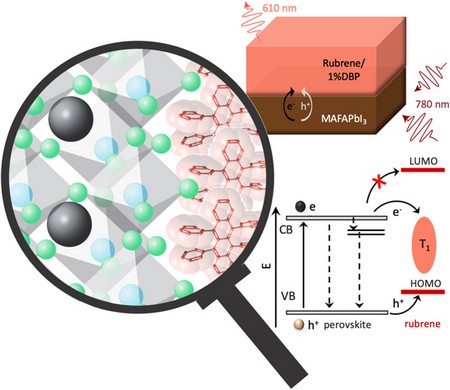
A perspective on triplet fusion upconversion: triplet sensitizers beyond quantum dots, MRS Communications

Triplet-pair spin signatures from macroscopically aligned heteroacenes in an oriented single crystal
The photoexcited triplet state of sapphyrin dication: Unusual spin alignment in monomers and spin delocalization in dimers - UNT Digital Library

Implementation of a Parallel Protein Structure Alignment Service on Cloud

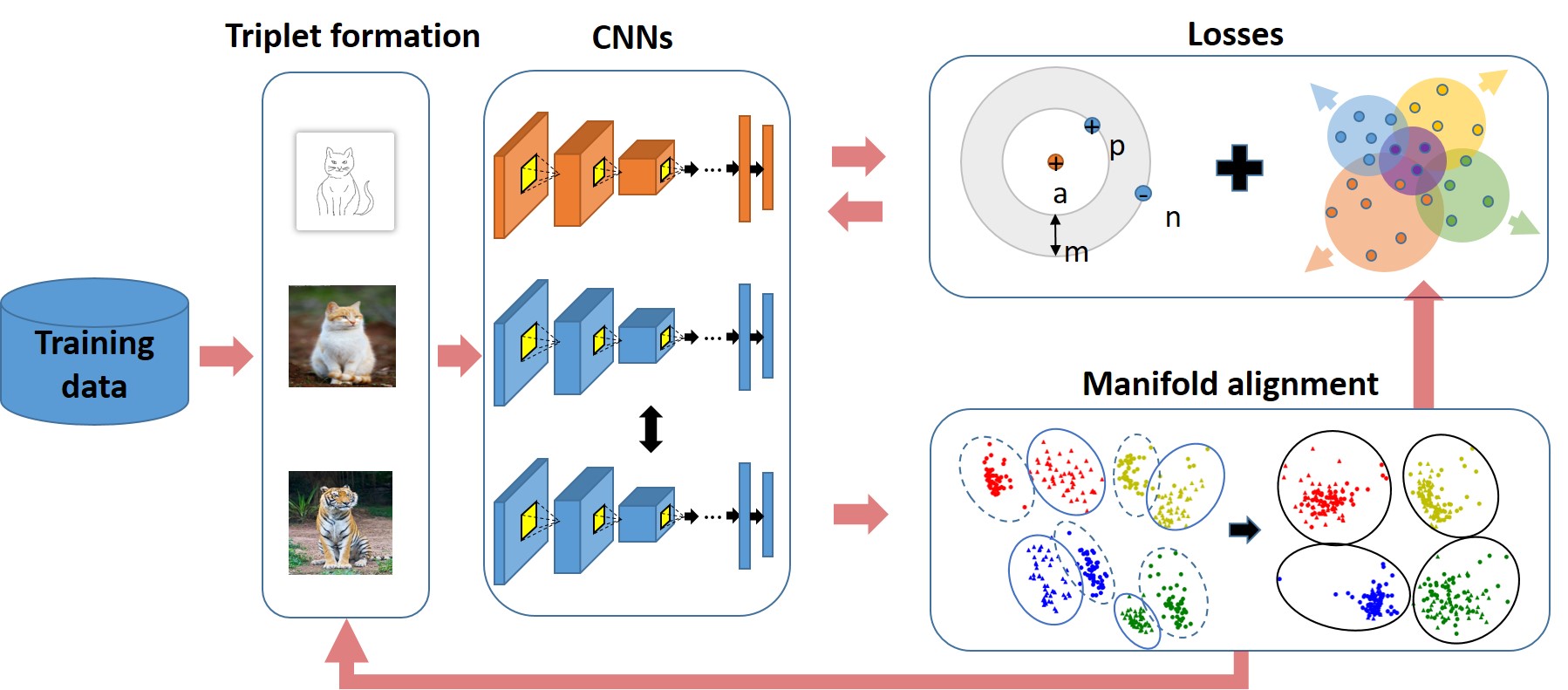
Joint Geometrical and Statistical Alignment Using Triplet Loss for Deep Domain Adaptation

Triplet-pair spin signatures from macroscopically aligned heteroacenes in an oriented single crystal

Triplet-pair spin signatures from macroscopically aligned heteroacenes in an oriented single crystal
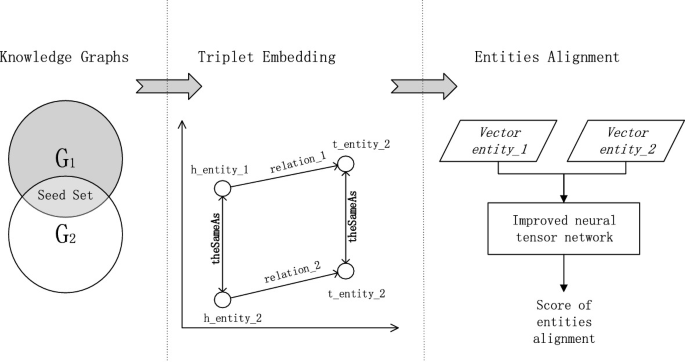
Cross-Knowledge Graph Entity Alignment via Neural Tensor Network
ACCV18
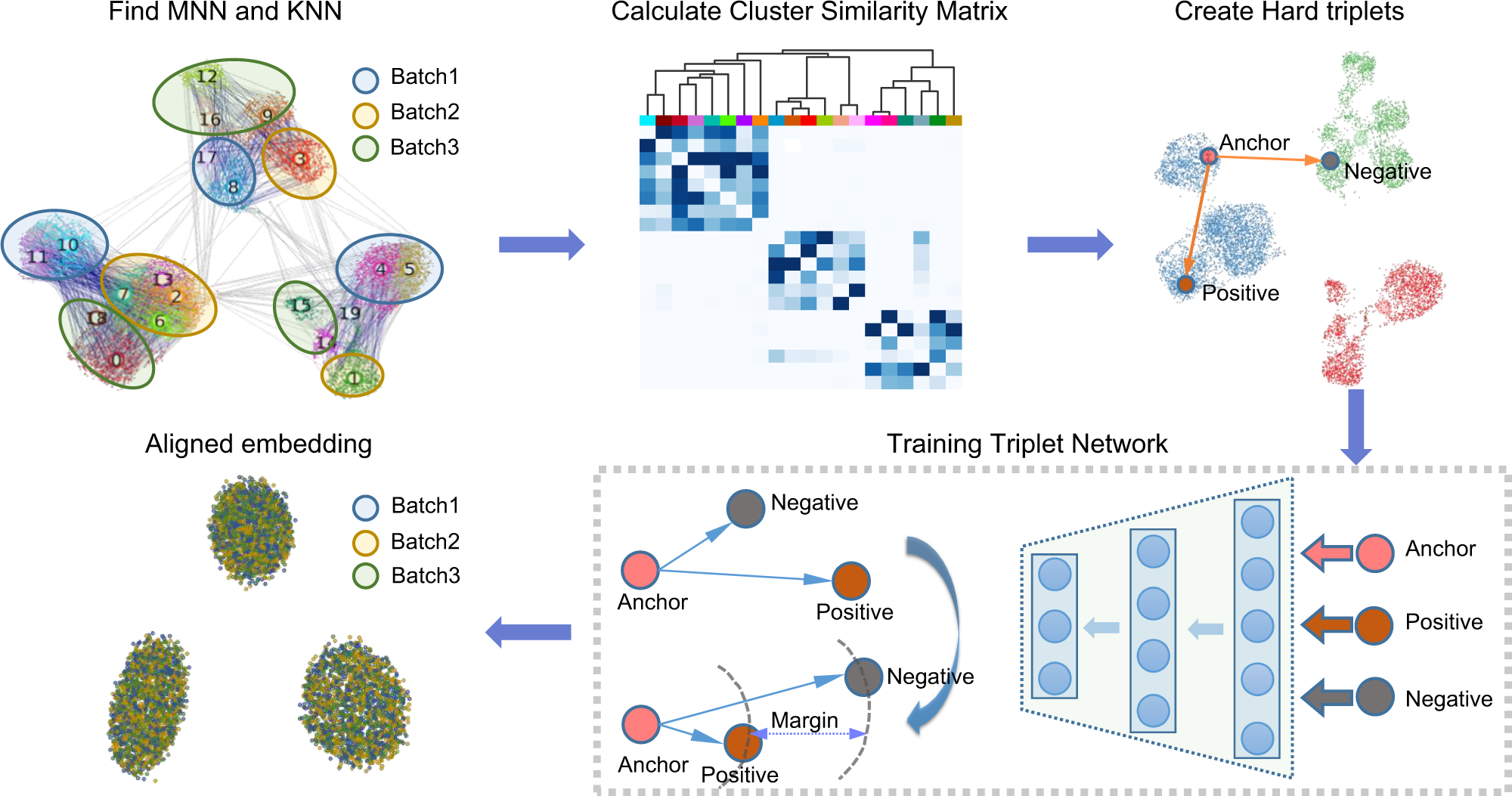
Batch alignment of single-cell transcriptomics data using deep metric learning
